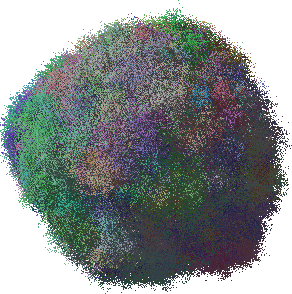TumourSimulator (V.1.2.3) - computer code for simulating solid tumours
This program simulates the model of tumour growth as described in
A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity,
Bartlomiej Waclaw, Ivana Bozic, Meredith E. Pittman, Ralph H. Hruban, Bert Vogelstein, and Martin A. Nowak,
Nature 525, no. 7568 (September 10, 2015): 261-64. doi:10.1038/nature14971
Please cite this paper in any work that has resulted from using this code.
All files can be downloaded as a ZIP file from here. Previous version 1.2.2 can be found here.
The program has been tested using GNU C++ compiler (g++) and Intel C++ compiler (icc). Assuming g++ is used, the program should be compiled as follows:
Compilation (Linux and Mac)
g++ simulation.cpp main.cpp functions.cpp -w -O3 -o cancer.exe
Compilation (Windows)
g++ simulation.cpp main.cpp functions.cpp -w -O3 -lpsapi -o cancer.exe
Usage
cancer.exe [name] [number of samples] [seed for the random number generator] (Windows)
./cancer.exe [name] [number of samples] [seed for the random number generator] (Linux and Mac)
where [name] is the name of a subfolder created by the program to which all data will be saved. For example, when executed with default parameters as
cancer.exe test 1 1 (Windows)
./cancer.exe test 1 1 (Linux and Mac)
the program generates output that can be downloaded here.
Parameters
All parameters are stored in the file params.h. Their names are self-explanatory, there are also comments in the file that explain the action of some switches (implemented using the compiler directive #define).
Interactive version
An interactive version of TumourSimulator can be found here (MS Windows only).
It opens a window which allows the user to set the parameters of the simulation, and set up video recording.
The software uses the same simulation algorithm as the command-line version described above, but some functions have been disabled and cannot be re-enabled without recompiling the code.
This program is intended for visualisations only.
Example code in Mathematica for generating images
The output of TumourSimulator can be used to generate three-dimensional images of tumours. Here is an example notebook written in Mathematica which
uses the data files generated by the above example ("test") to produce a 3d tumour with cells colour-coded in a way similar to that from Fig. 2 from the paper:
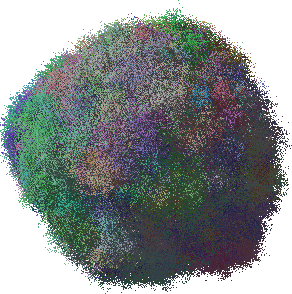
This example shows how to make a 2d slice through the centre of the tumour. Using the same data as for the above 3d visualisation, the Mathematica notebook shows how to label three most common mutations with red, green, and blue (left picture) or how to label cell lineages (right picture) so that cells with similar PMs obtain similar colours.


Copyright and licensing
Copyright (2015) The University of Edinburgh. TumourSimulator is free software: you can redistribute it and/or modify
it under the terms of the GNU General Public License as published by
the Free Software Foundation, either version 3 of the License, or
(at your option) any later version.
TumourSimulator is distributed in the hope that it will be useful,
but WITHOUT ANY WARRANTY; without even the implied warranty of
MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE.
See the GNU General Public License for more details.