Current research interests
[A] Self-assembly & MECHANICAL PROPERTIES virusES
[B] The biophysical Basis for bacterial Life & Death
[C] pickering emulsions & microfluidics
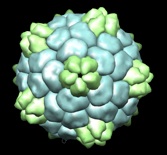
We use neutron and x-ray scattering in combination with lab-based methods to learn how viruses self-assemble, can be triggered to disassemble and exhibit the mechanical properties needed to survive. In collaboration with the Young group, we have performed yeast expression of the capsid proteins we require for these experiments. We have also been fortunate enough to work with Prof. George Lomonossoff of the John Innes Centre.
As part of the Length scale bridging in biophysical systems program that has been established with the NPL, we are using a combination of neutron reflectometry from floating bilayers and microbial studies using optical microscopy to learn how anti-microbial and cell division peptides interact with bacterial membranes.
We have developed gold nanoparticles with pH- & temperature-responsive grafted polymer coatings as stabilizers for Pickering emulsions. We are now using these stabilizers in combination with microfluidic approaches to droplet production, to produce monodisperse emulsions. We are using these droplets to conduct highly parallelized experiments to gain insight into projects [A] & [B] above.

20μm

Chloroform (nile red stain) in water PDMAEMA-functionalized gold nanoparticle-stabilized Pickering emulsion at pH 2 produced by shaking and extrusion.

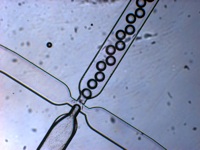
Emulsion droplets produced by microfluidics. (Stuart Conley)