Welcome to the Topologically Active Polymers (TAP) Lab!

Group mission:
In spite of their extreme length and confinement, our genomes are surprisingly well organised, functional and knot-free. This is achieved via sophisticated proteins that exert exquisite topological and mechanical control over the genome's material properties.Inspired by this, we aim to discover new DNA-based topological soft materials and complex fluids that can change properties in time.
The group's expertise is rooted in polymer and statistical physics, and employ both simulations and experiments to answer our questions.
We believe boundaries between disciplines were made to be broken, and we do our best to shatter them every day.
News:
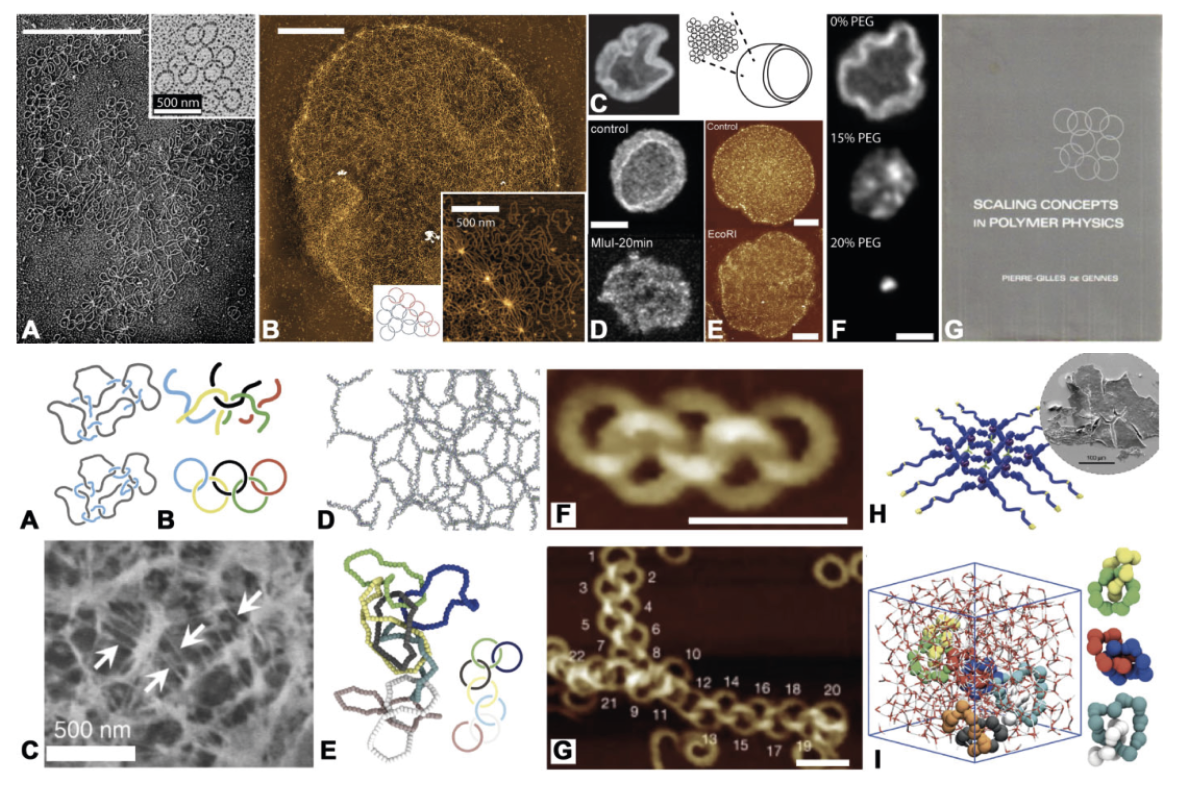
1/2025 Big paper out! - all credits to Yair and Giorgia who led this tirelessly until the end! We discover that DNA nanostars form gels with microscopic interlinked loops that determine the macroscale elasticity of the gel! The story is now out in Nature Materials!
Link to paper
Link to News and Views "When Stars make Loopy Networks" by Emanuela del Gado
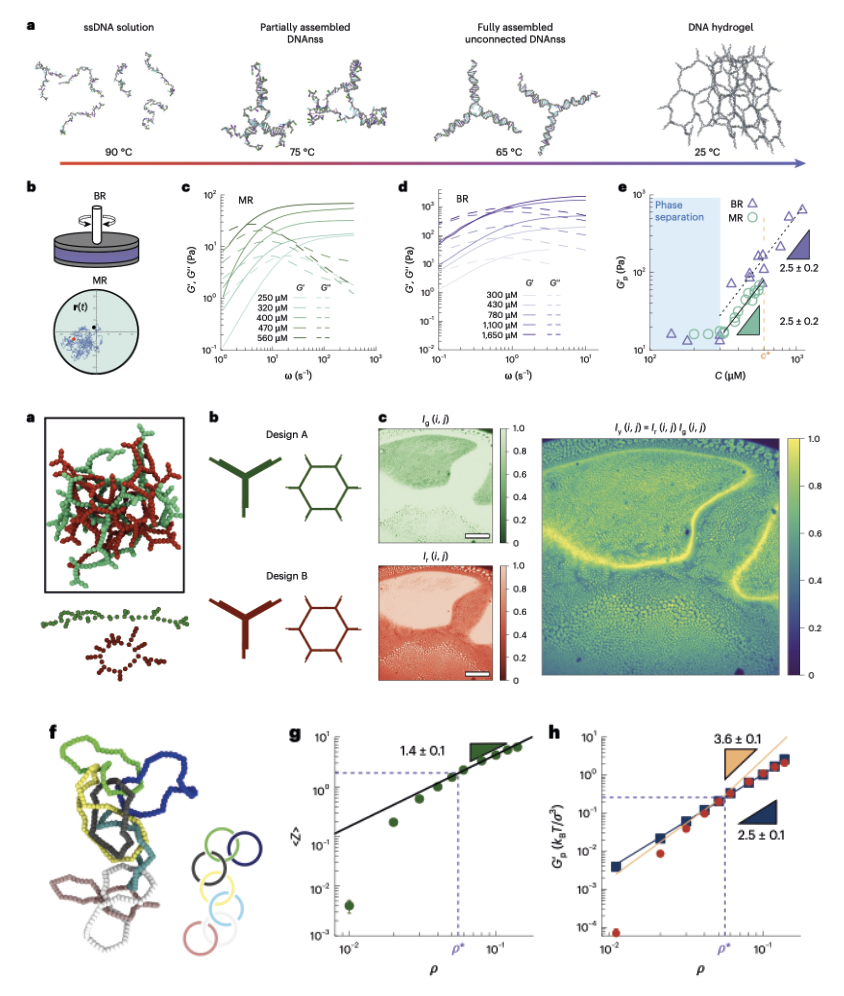
1/2025 Kinetoplast DNA: A polymer physicist's topological Olympic dream. A review on kinetoplast DNA and comparison with DNA nanostructures our in NAR!
Link to NAR article
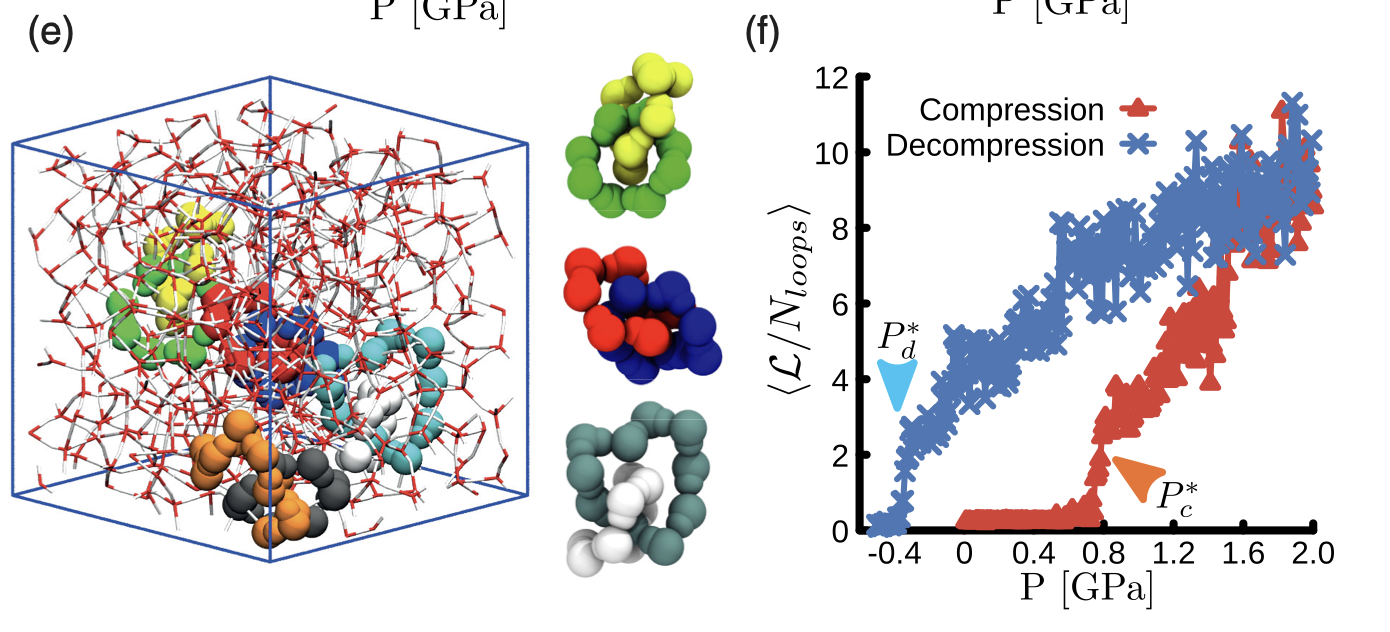
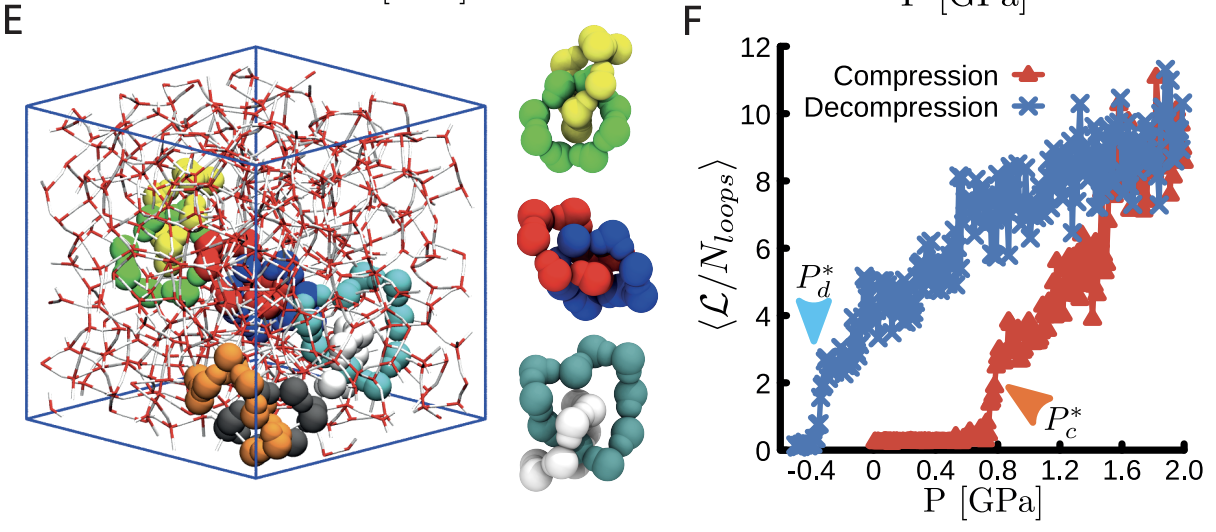
12/2024 Link to Densify! How water molecules tie themsleves into knots to form high density amorphous ice, now published in PRL!
Link to PRL article
12/2024 Our Physics World article "Spot-The-Knot" is live!
Spot the Knot article on Physics World
Link to spot-the-knot game by Djordje

10/2024 So happy to have been awarded the 2024 Leverhulme Prize in Phyisics!
This award is given to 5 physicists every 3 years across all disciplines, so honoured to be one of them!
Link to Award
Link to Wikipedia

09/2024 Thank you British Biophysical Society for the Louise Johnson Early Career Award! I will give my award lecture at the biannial BBS meeting in Swansea (11 September).
Link


09/2024 Welcome Djordje, Sabrina and Lirong. They will work on ML of knots, DNA in extreme conditions and kDNA!



06/2024 We published on water!
Link to Densify: Topological Transition During the Compression of Amorphous Ices
03/2024 The kinetoplast DNA is a gift that keeps giving! Another work from the group, led by Sam, on how to destroy kDNA using restriction enzymes one minicircle class at a time!
Pulbished in PRX Life
01/2024 Welcome Auro and Emma in the team. They will work on drug discovery - a new adventure of the TAPLab!


01/2024 Check out this RSC blog from myself on "career-related advice" and "the most important questions to be asked in our field". I tried to be thought-provoking! I'd be delighted to hear what you think! Have a read at this link!
01/2024 Great start of the year with the first 2024 cover of Soft Matter!
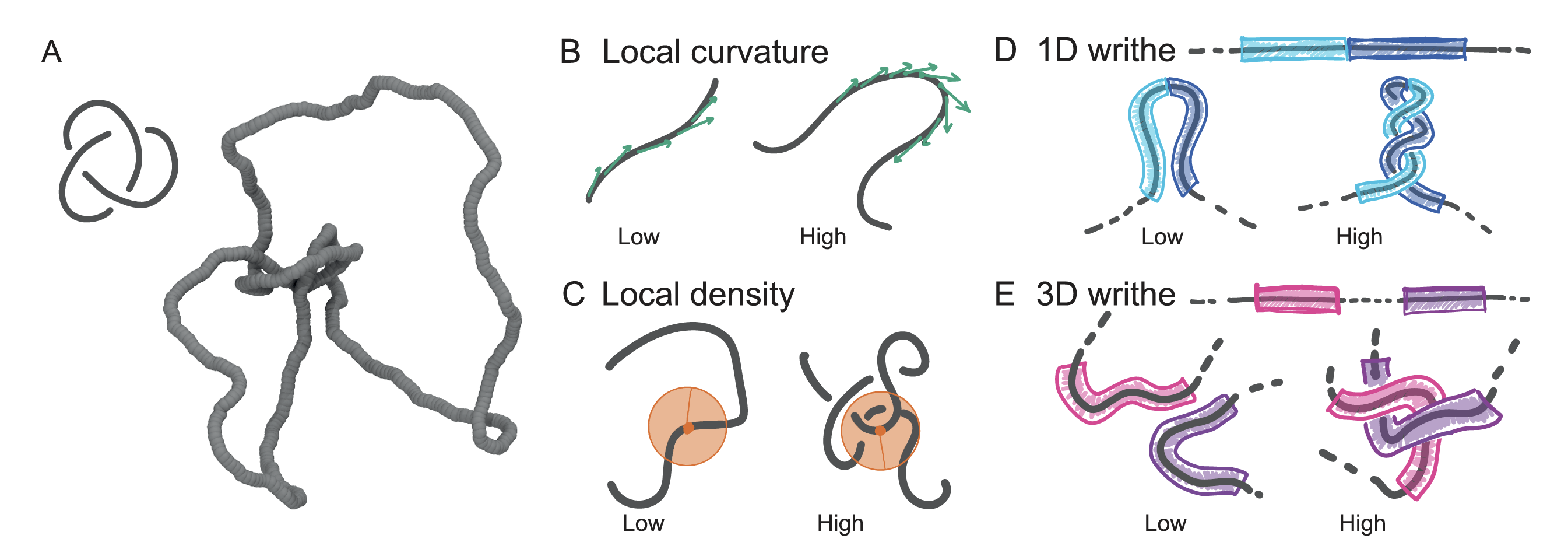
Spearheaded by Joseph and Filippo, we created a database with around a million knotted configurations up to 10-crossings and trained a ML to identify them.
Turns out that a "local writhe" descriptor yields the best accuracy, up to 95% for 250 different knots!
Read more at this Link!

07/2023 Well done Yair for making the cover of Soft Matter with an impressive paper on the rheology of DNA nanostars! Read more at this Link!

05/2023 The "Topological Soft Matter" workshop in Edinburgh was a lot of fun! So much amazing science and such a friendly and open atmosphere. Hope to run it again. Below the recording of my talk!
04/2023 Welcome to Ella! The first PhD student to venture in the realm of polyrotaxanes!

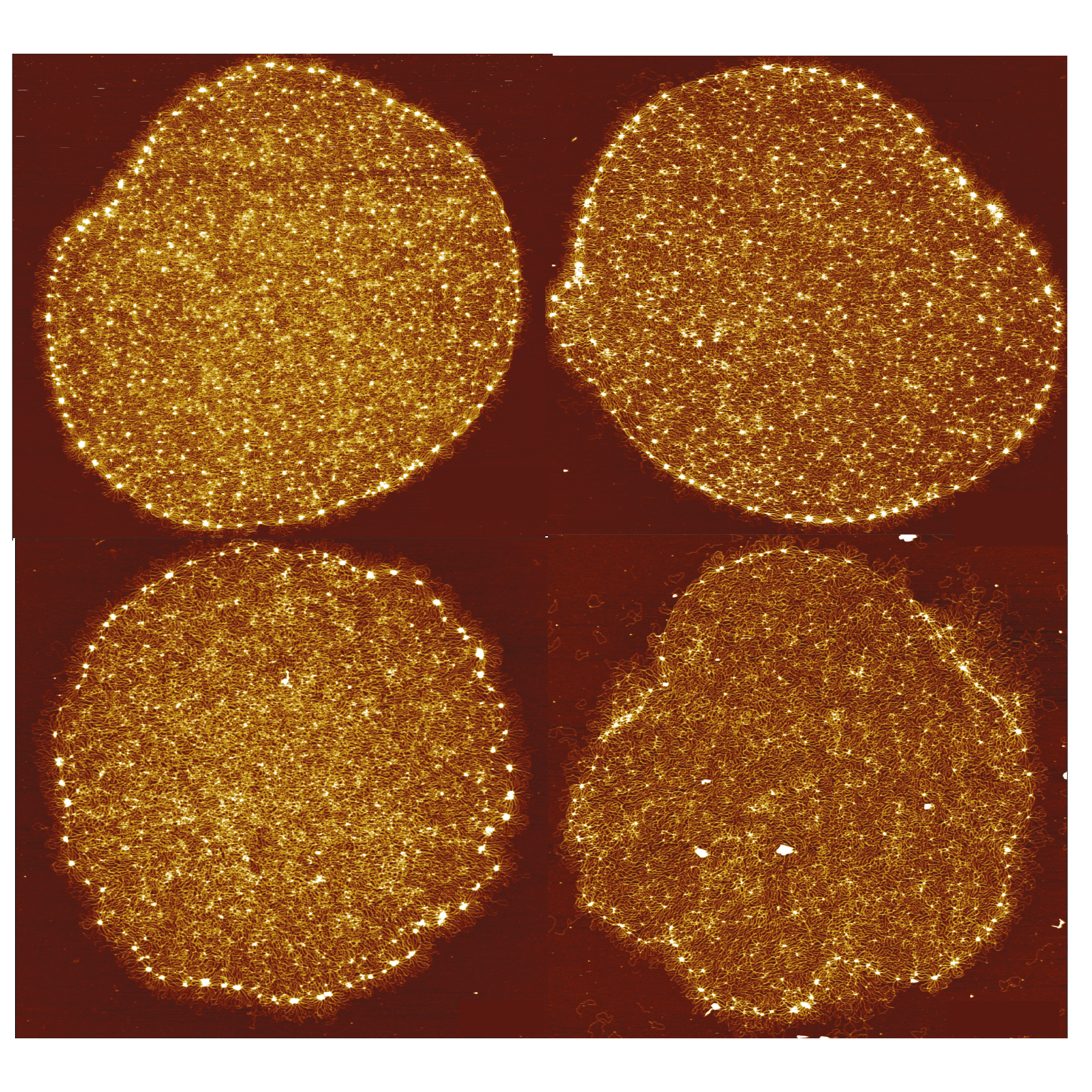
04/2023 So many beautiful pictures! kDNA will never stop to amaze. We have used high resolution AFM and AFM-steered simulations to reconstruct the structure and topology of kDNA at single molecule resolution. Now open access in PRX!

The code for the AFM-steered MD simulations is also open access at
TAPLab gitLab!
01/2023 Excellent start of 2023 - Yair's paper on fluidification of DNA entanglements by IHF protein is published in PRL!
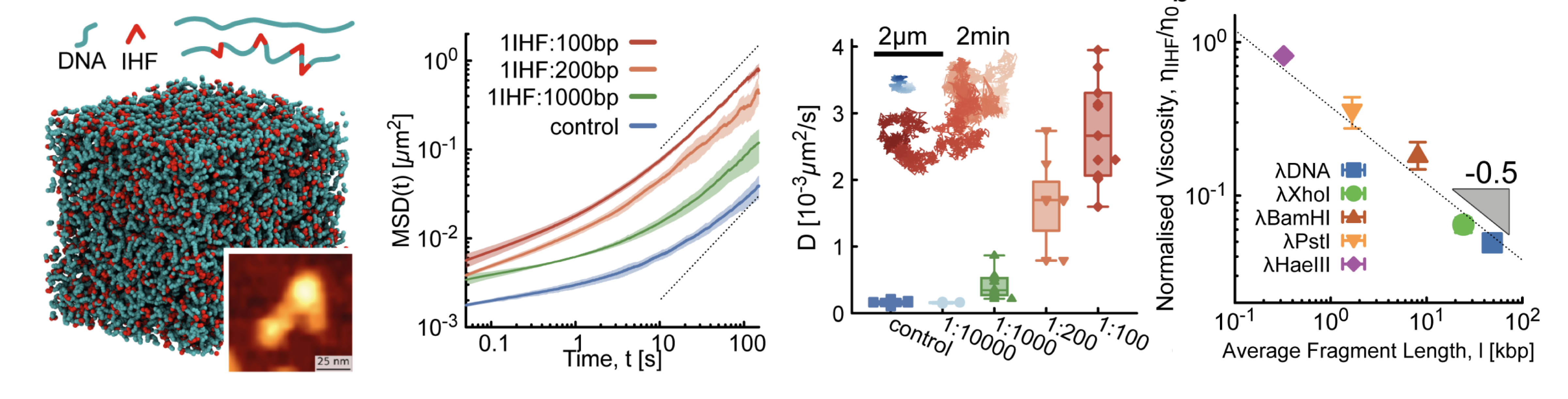
Related short talk here
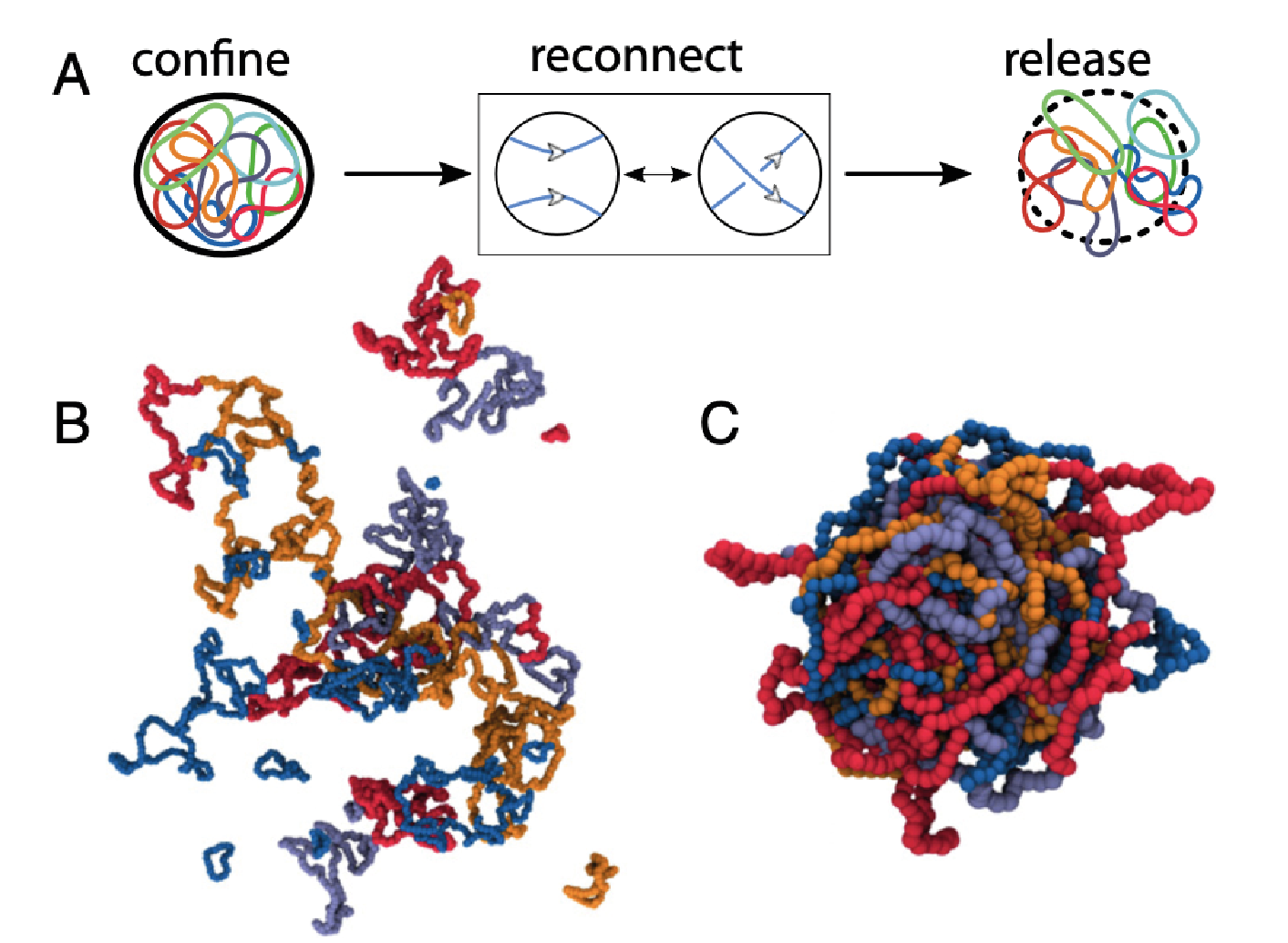
10/2022 Very cool paper by Andrea Bonato, who shows that reconnecting ring polymers under confinement can undergo "topological gelation", forming knots and links. Read more in PNAS.
11/2022 Welcome to Dinesh, Filippo and Sam who join the TAPLab!



08/2022 Here we show that ParB from Bacillus subtilis can recruit other ParBs and load them onto DNA away from parS! Using simulations we show that this cooperative recruitment can lead to spreading along DNA. Experiments done by Milos Tisma and simulations from the TAPLab. Read more in Science Advances.
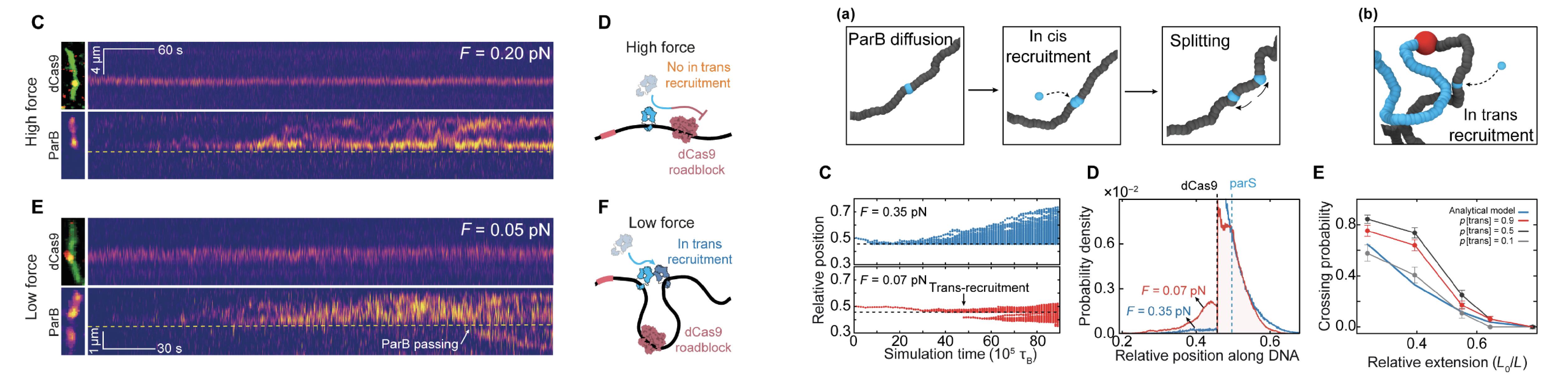 Movie showing the low force regime
Movie showing the low force regime
Movie showing the high force regime
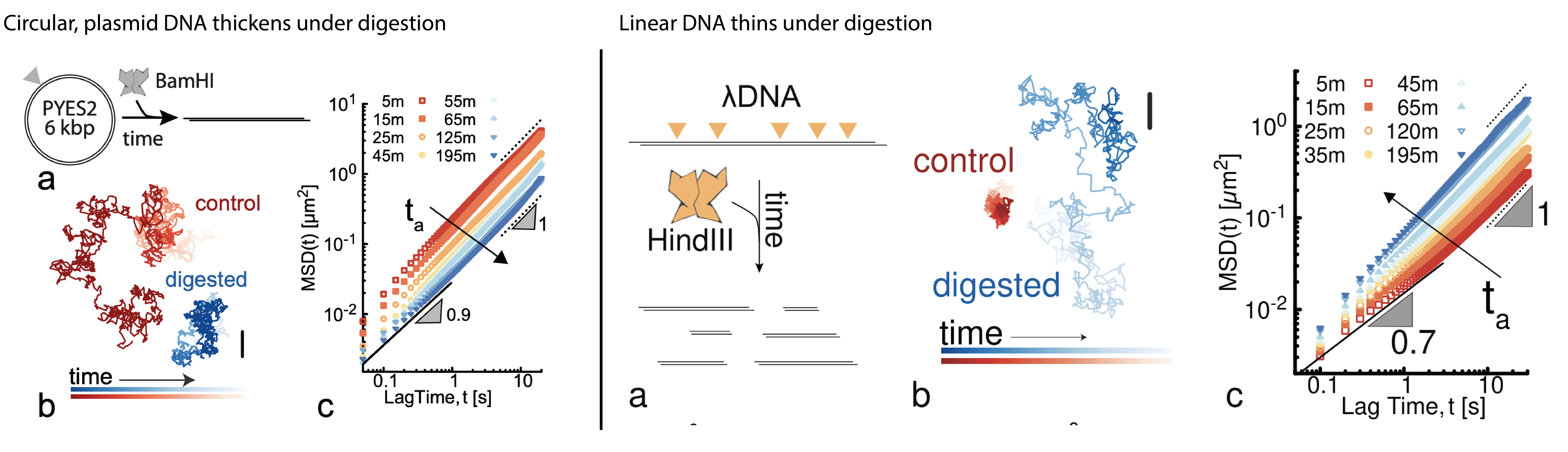
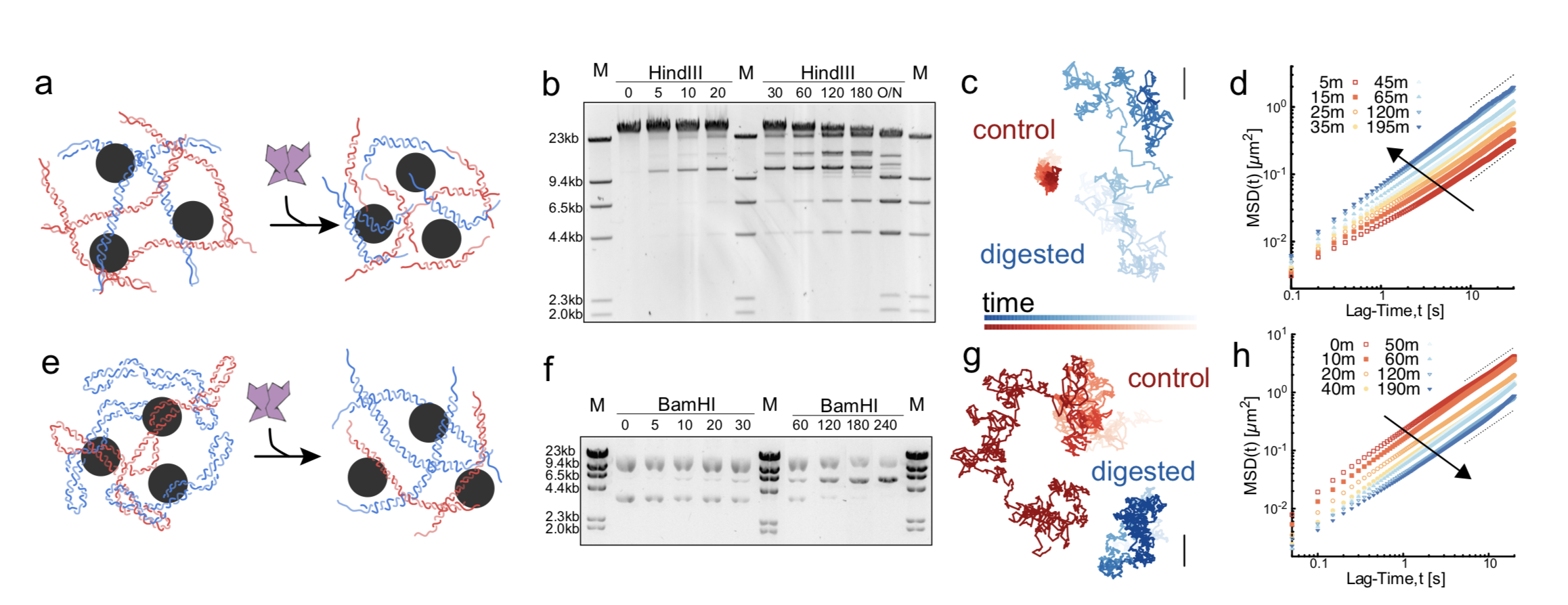
08/2022 In this paper we show that digestion of dense solutions of DNA yields interesting rheological signatures, and that they depend on the topology of the DNA! Read more in Nature Communications.
07/2022 New paper! We asked if there exist some local geometric properties of 3D curves that capture their "knottedness". Indeed there are! Patterns of local density and writhe can idetify the loction of knots along curves around 90% of the times! Read more here in ACS Polymers Au.
07/2022 A 12 minutes recorded talk from the APS March meeting on how IHF proteins can fluidify entangled solutions of DNA.
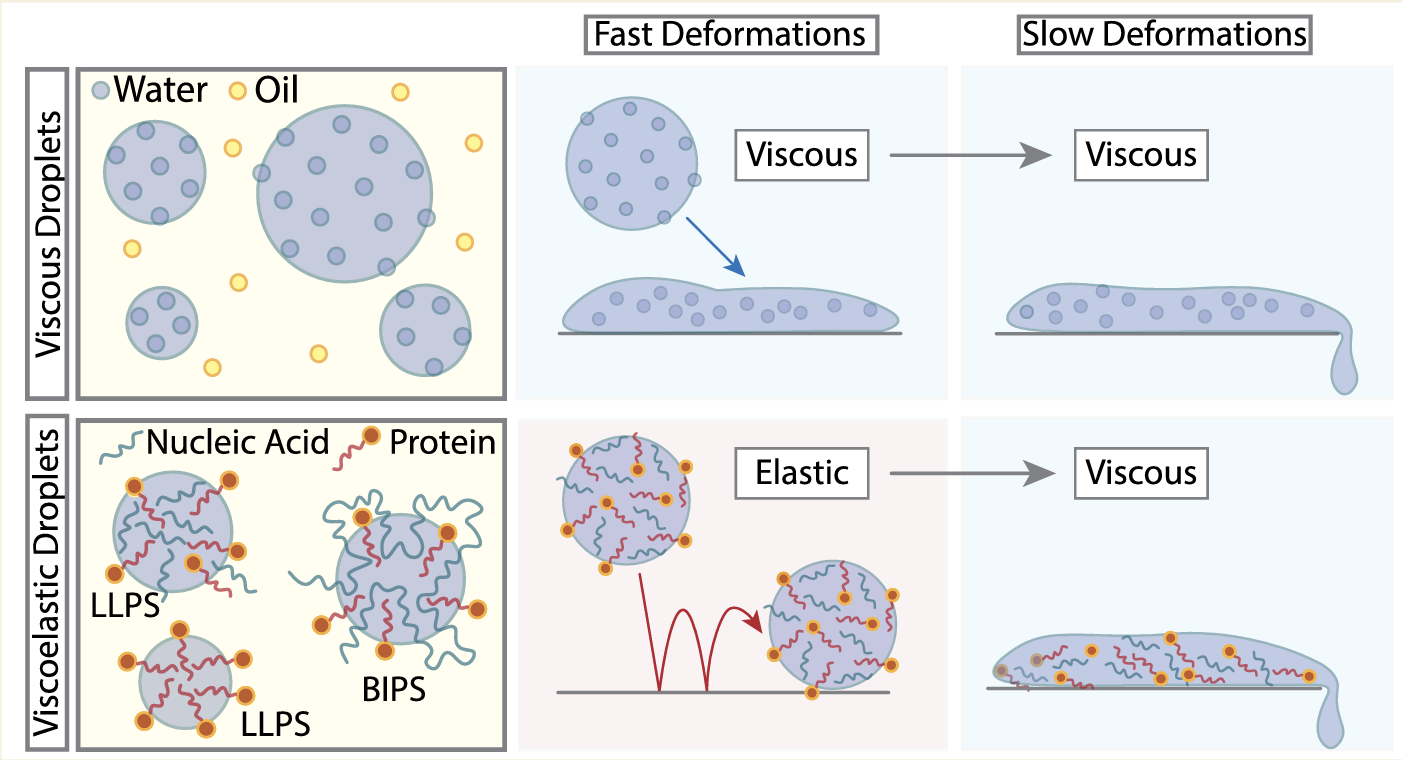
06/2022 New paper from the Lab! This time a perspective on the Viscoelasticity of Proteins and Nucleic Acids Condensates, written with Mattia Marenda!
Take home message: condensates are "by default" not liquid but viscoelastic. We review how to best measure their viscoelasticty and what could be the physiological implications of non-Newtonian rheology.
Now published in JACS Au.
05/2022 Really enjoyed Pint of Science 2022. I think I benefitted more than the audience - inspiring questions and fresh ideas! I loved it! You can see the video here, apologies that it was taken from a very tight angle!
04/2022 Welcome to Amandine, who joins the lab for her PhD on viscoelasticity of DNA-NANOG solutions!

04/2022 Physics of Life Summer School under way!
120 people, 5 days, 2 poster sessions, 2 Q&A sessions, excellent speakers and science, lots of ideas and infinite energy!


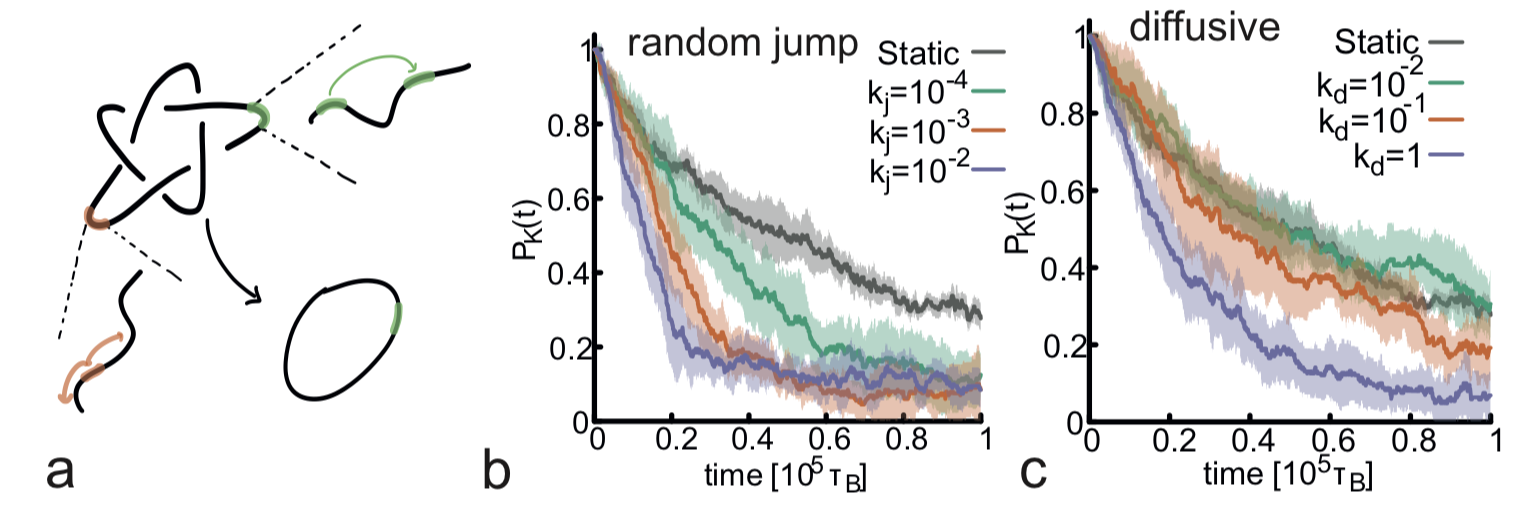
04/2022 Fresh publication in NAR. What's the most efficient way to keep things unknotted? Jump around!! Credits to Yair A. G. Fosado for most of the simulations! See GitHub for open source codes.
Dynamic and Facilitated Binding of Topoisomerase Accelerates Topological Relaxation
12/2021 New paper in collaboration with Dekker's lab
Condensin extrudes DNA loops in steps up to hundreds of base pairs that are generated by ATP binding events. Simulations again by Andrea, the Cook, Bonato.
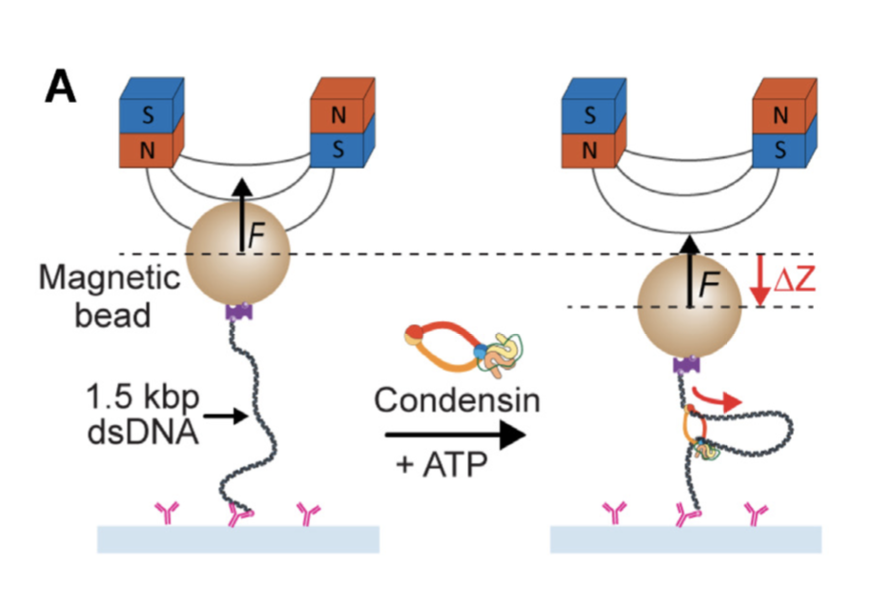
12/2021 Loop Extrusion goes 3D! Read more at
Biophys J.. Simulations by Andrea, the Cook, Bonato.
11/2021 TAPLab having a lasagne-off competition! The conclusion of the party was that if we don't have a future in science we can set up a rather decent osteria and sell food!



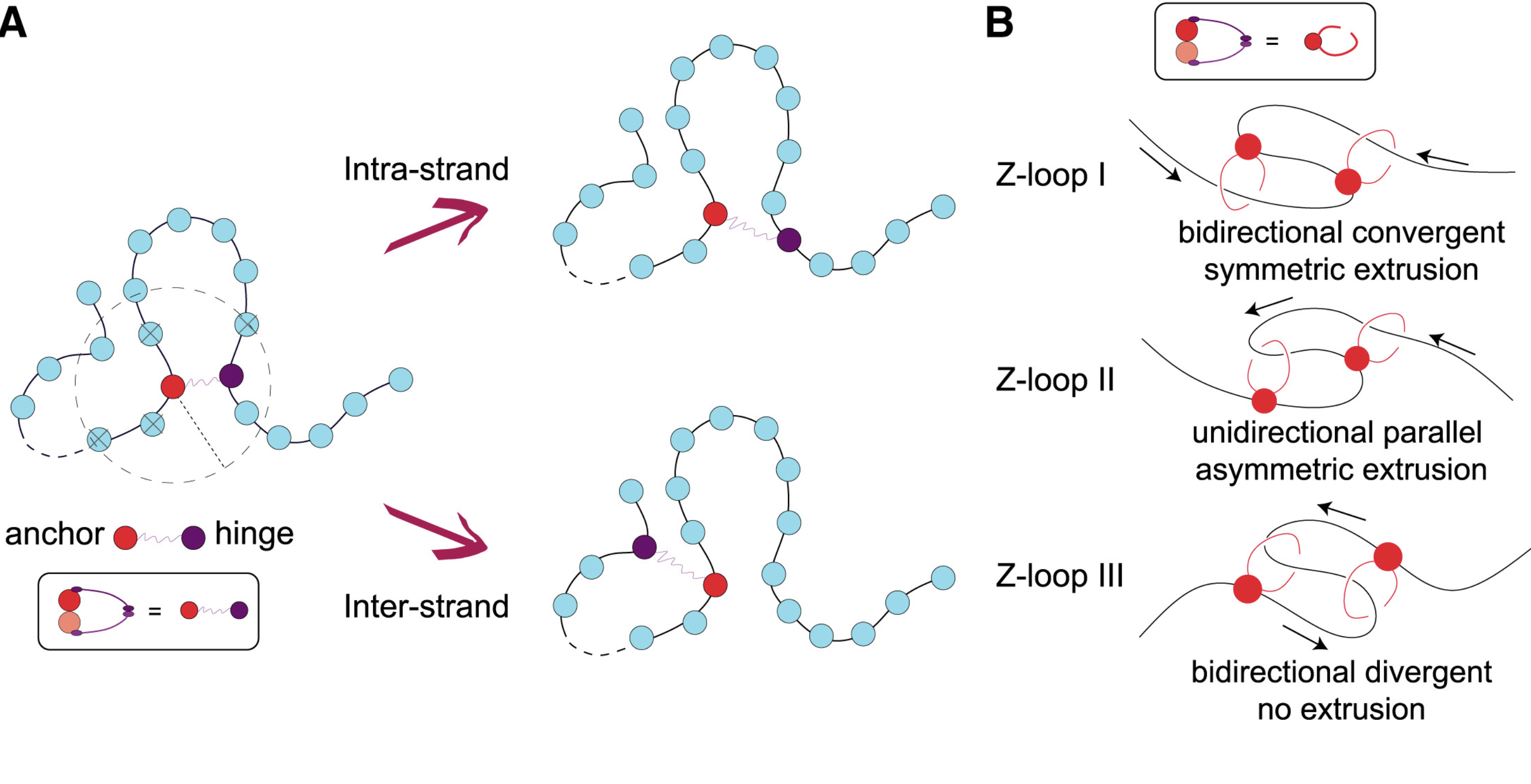
10/2021 Entropic Competition - who's going to win? Read more at PRR
Simplifying topological entanglements by entropic competition of slip-links. Simulations again by Andrea, the Cook, Bonato.
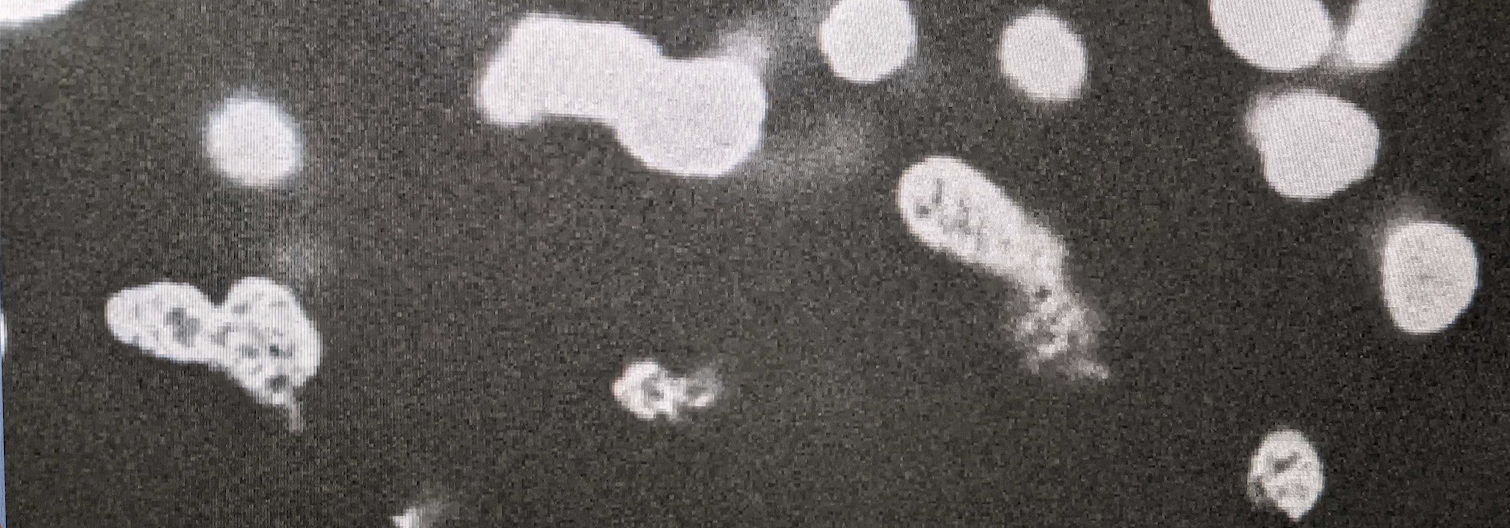
10/2021 Ah! Our friend the Kinetoplast DNA imaged with a spinning disk confocal - thanks to the magic hands of Mattia Marenda at IGMM and ESRIC

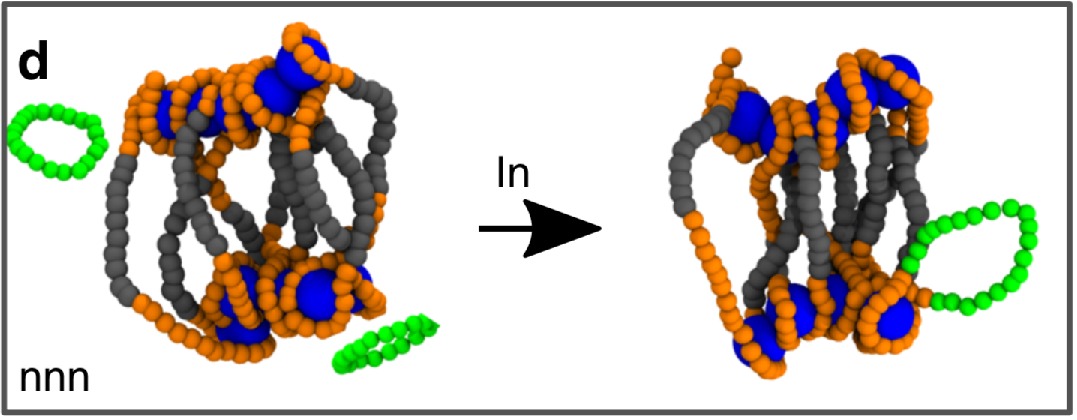
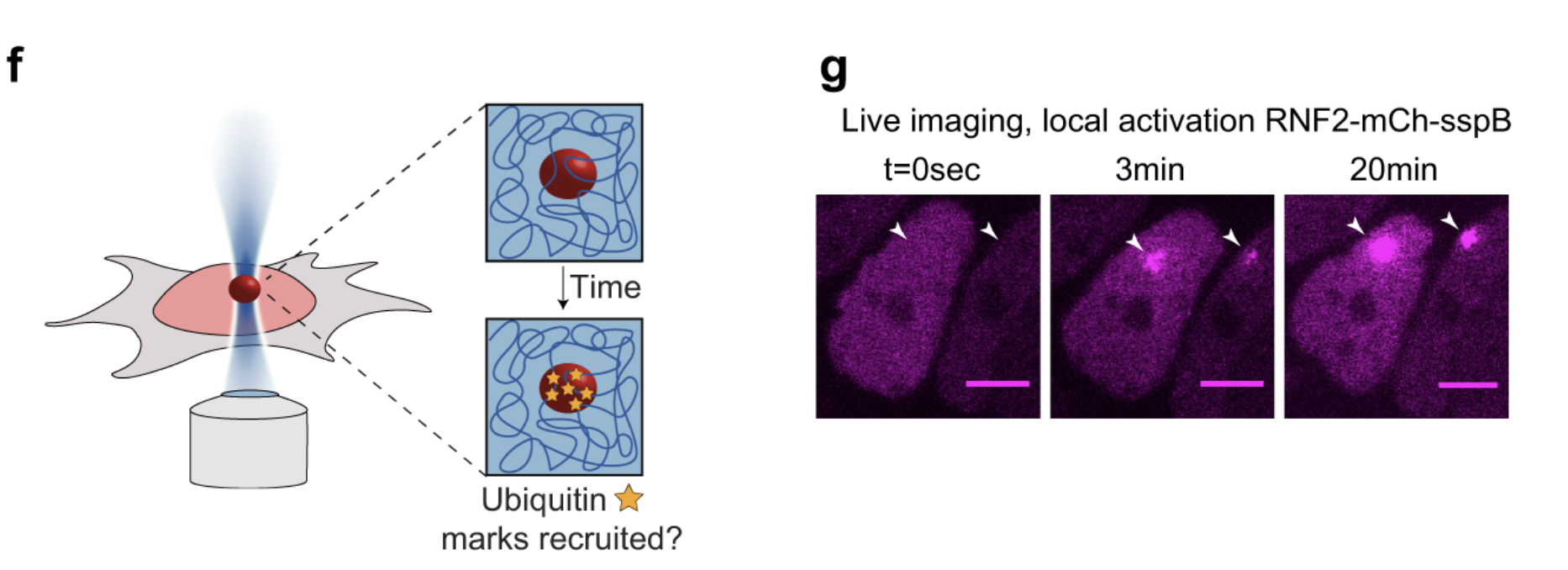
10/2021 New paper on epigenetic memory and "recolourable" polymers in Nature Communications.
Polycomb condensates can promote epigenetic marks but are not required for sustained chromatin compaction All credits to Jorine Eeftens while in Brangwynne lab. Using optogenetics she showed that PRC condensates can compact chromatin but that they are are not needed to maintain the compact state. A very interesting link between thermodynamics and epigenetic memory!
08/2021 New preprint! By mixing entangled solutions of DNA with restriction enzymes we discover that we can obtain both thinning and thickening! Find out more at
Universal and Programmable Rheology of Topologically-Active DNA Fluids
05/2021 Finally out - Topological tuning of DNA mobility in entangled solutions of supercoiled plasmids in Science Advances!
05/2021 Maria and Yair join the lab!
Maria will be the expert on living polymer simulations while Yair will model DNA hydrogels and origami in their finest details!


05/2021 Giorgia looks so well after her DNA hydrogels that they sometimes show their affection back ..
04/2021 Jenny joins the lab, she will be working on rheology of DNA origami!

04/2021 An exceptionally talented MPhys student Liam Bradbury (Link to fb) made this beutiful public summary on his MPhys project, a must watch and so catchy!
03/2021 What can soft matter do for biology (and viceversa)?
Mattia Marenda (IGMM) and Davide try to convince molecular biologist that soft matter is cool!
03/2021 Fun (or embarassingly unprepared) Higgs chat w Davide.
The associated Higgs talk was far better prepared (but ran long) on topological glasses, magnetic polymer models for chromatin and Progressive DBSCAN clustering (= a random bunch!).
03/2021 Talk on HIV integration, topological regulation of the genome and TAPs at GEOTOP-A
03/2021 Giorgia joins the lab, a chemist by training, she will be working on DNA hydrogels!

02/2021 New paper from Yair and Davide Non Equilibrium Supercoiling and Action at a Distance now out in PNAS!
02/2021 Davide receives the Early Career Award in Statistical Mechanics and Thermodynamics from Royal Society of Chemistry!
02/2021 New paper from the group! Andrea Bonato and Davide collaborate with Cees Dekker lab to explain phase separation of yeast cohesin in vitro. See Bridging-induced phase separation induced by cohesin SMC protein complexes in Science Advances!
01/2021 Cleis, Ed and Simon join the lab! Cleis will be working on models of genome organisation, HIV integration and transposable elements, Ed on material properties of genomes, kinetoplast and the sorts while Simon will bring in some proper experimental (micro)rheology expertise.


11/2020 Davide receives the Chancellor's Rising Star Award from the Univeristy of Edinburgh!
10/2020 Non-Equilibrium Living Polymers. A fun exercise in scaling calculations! Open access in Entropy (invited by A Maritan and A Giometto) Link
09/2020 Finally I can say it (embargo lifted): ERC StG grant awarded! Hurray! Link See Team&Vacancies for openings in the lab
08/2020 Another grant awarded (can't say which yet)! Keep an eye out for more available positions in the lab!
06/2020 I am very honoured to have been awarded a Royal Society University Research Fellowship! This adventure including experiments on DNA topology and microrheology will start in Jan 2021. A PhD position will be soon advertised (drop me a line if interested!).
05/2020 Very happy to have been nominated "Outstanding Reviewer" by RSC Soft Matter!! href="https://pubs.rsc.org/en/content/articlelanding/2020/sm/d0sm90069a#!divAbstract">Link
05/2020 Threading induced dynamical transition in Chimeric Polymers out in ACS MacroLetters. Great collaboration with Angelo Rosa, Jan Smrek and Matthew Turner also part of the EUTOPIA COST action.
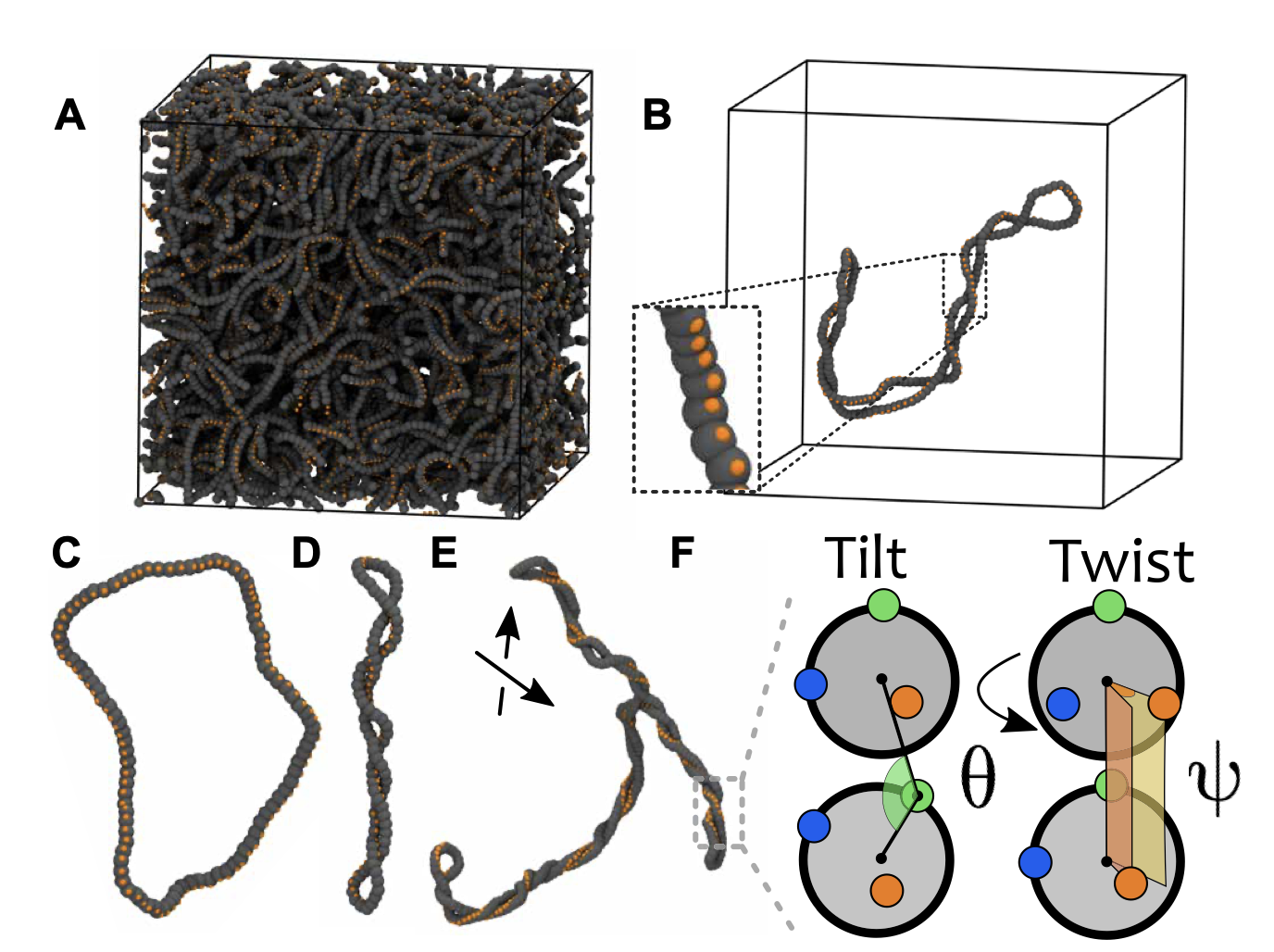
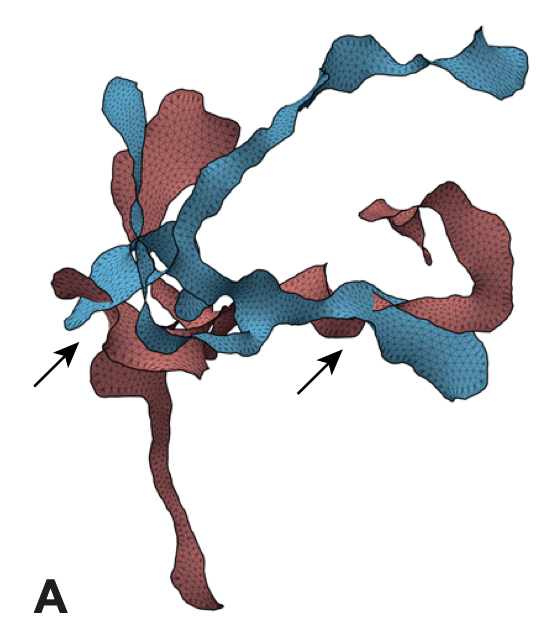
04/2020 Beautiful representation of the action of SMC and Topoisomerase on knotted strings! Courtesy of Elias Melas 03/2020 In May I will start a Leverhulme Early Career Fellowship at UoE on "Topologically Active Polymers" (TAPs)! I will do (try) theory and experiments so need lots of help. Drop me an email if interested to work on this!
02/2020 Great collaboration with Alex Bousios and Hans-Wilhelm Nuetzmann on the interplay between Transposable Elements and 3D genome organisation now out on Mobile DNA
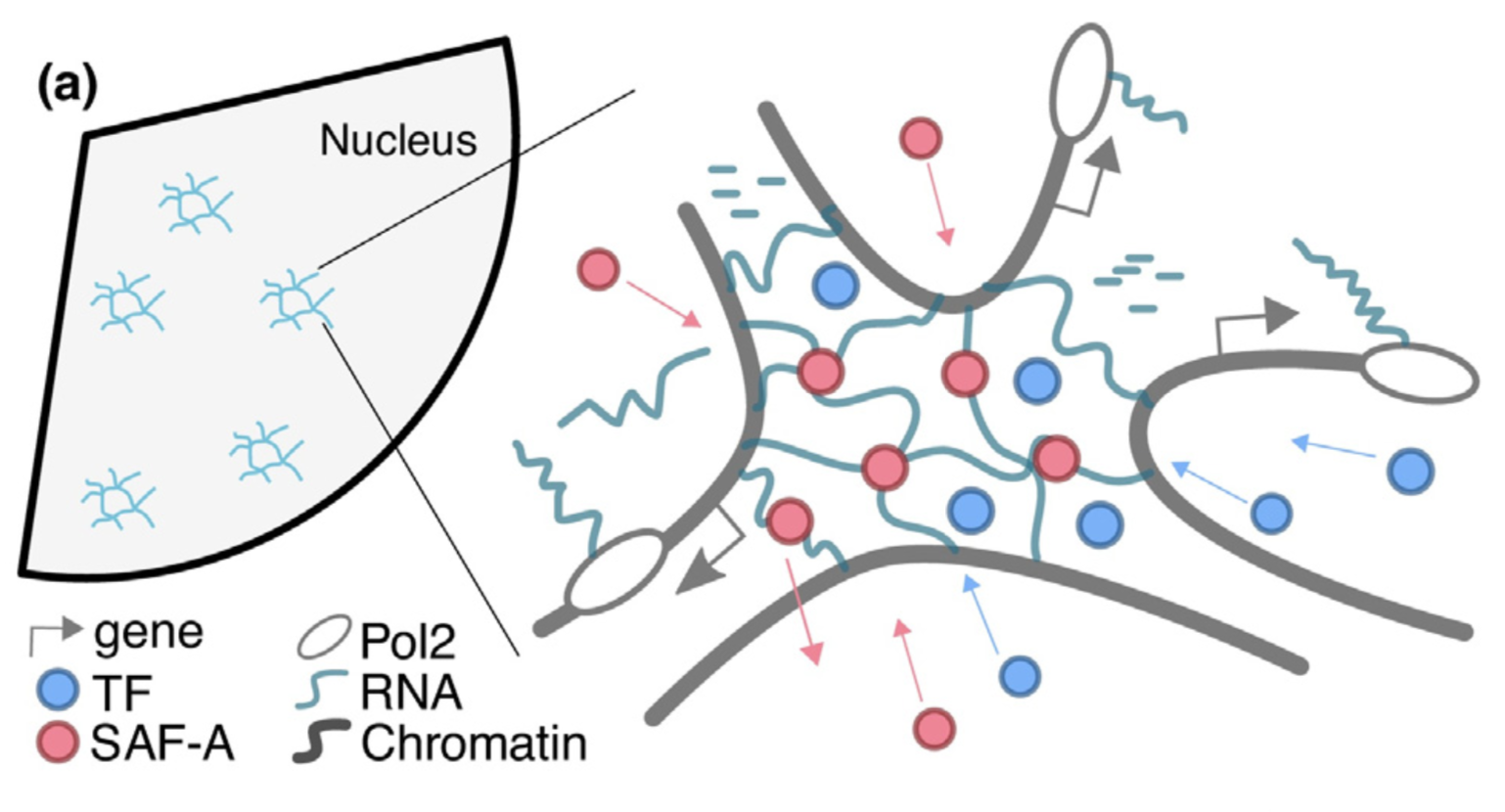
05/12/2019 Welcome Gaia! 09/2019 Non Equilibrium Theory of Epigenomic Microphase Separation in the Cell Nucleus 08/2019 Looking forward to Physics Meets Biology 2019 in Oxford in September! Link
08/2019 Very proud to have been awarded a Leverhulme Early Career Fellowship which will start in May 2020 at the University of Edinburgh 08/2019 Excited to start my new position as Research Fellow in the Dept. of Mathematical Sciences in Bath! 05/2019 Meeting on 4D genome organisation in Venice (3-5 October 2019): More informations and details HERE and yes, the venue of the conference is the one in the picture! :) 05/2019 Very productive meeting with Hans Neutzmann and Alex Bousios on the topology and topography of transposable elements! Many thanks to the Physics of Life Network for funding! 04/2019 New review on the role of RNA on chromatin organisation 04/2019 Interdisciplinary Challenges in Non-Equilibrium Physics going at full steam!
03/2019 "Synergistic Topological Simplification of the Genome" now in PNAS 01/2019 "Physical Principles of Retroviral Integration" now in Nature Communications 11/2018 With the support of the Higgs Centre, the Physics of Life and the Emergence and Physics Far From Equilibrium Networks George Constable (Bath), Gianmaria Falasco (Luxembourg), Elsen Tjhung (Cambridge) and I will be hosting an "Interdisciplinary Challenges in Non-Equilibrium Physics" Workshop in Edinburgh (10-13 April 2019, LINK).
05/2018 Congratulations to Luca Tubiana and Raffaello Potestio for organising and winning a COST action on Topology. EUTOPIA will connect people working on a range of topics, from liquid crystals to chromatin! A great chance for new collaborations and projects!!
01/2018 The first workshop on the Physics of Epigenetic and Chromatin Dynamics will be held in Edinburgh 16-17 April 2018. The registration is free but only a limited number of places are available. For poster abstract submission please email davide.michieletto@ed.ac.uk. 01/2018 Our recent paper on Epigenetic Memory and Genomic Bookmarking has been published in Nucleic Acids Research!
11/2017 Paper in collaboration with Angelo Rosa on the glassy dynamics of ring polymers under random pinning has been published in PRL
09/2017 Congratulations to Chris for his paper on Non-Equilibrium Chromosome Looping. An alternative view to loop extrusion "without a motor"! Made the cover of the Sept. issue of PRL!! 20/05/2017 Welcome Dante!
12/2016 Our paper on a Recolourable Polymer model to describe chromatin folding with an underlying dynamic epigenome has been published in PRX, followed up by an APS Focus story on How Cells Remember Who They Are signed by science writer Philip Ball
12/2016 My paper on the Tree-Like Structure of Rings has been published in Soft Matter! 05/2016 Last paper of my PhD: A Topologically-Driven Glass in Ring Polymers has been published in PNAS with a News&Views in Nature Physics!
10/2015 Paper on the Topological Patterns of DNA Knots accepted in PNAS!!
09/2015 Very honored to receive the IOP Ian Macmillan Ward Prize for best PhD student publication in Polymer Physics
05/2015 My model for the Kinetoplast DNA a genomic "medieval chainmail" has been published in Physical Biology
12/2014 A Taste for Anelloni has been featured in the IOP magazine Physics World followed up by a new entry in the Wiktionary!



see also PNAS article.
out in PRL and companion PRE